
- 09 Nov 2015
This story forms part of a series of interviews highlighting the work of experts in the field of complex systems science. Dr. Sam Scarpino is an Omidyar Fellow at the Santa Fe Institute and an alumnus of the Santa Fe Institute’s 2010 Complex Systems Summer School. He will soon be joining the University of Vermont as Assistant Professor of Mathematics and Statistics.
Sam earned a B.Sc. in biology from Indiana University Bloomington and a Ph.D. in integrative biology from The University of Texas at Austin. His scientific research statement is interesting and diverse. Sam investigates pressing biological questions by integrating mathematical models and data with powerful statistical methods.
Q: You have worked on a very broad range of research questions - can you give us some examples of research you have been involved in?
The diversity of topics I’ve worked on largely derives from the fact that I started off as a biologist that worked in the field and in the lab, doing a little mathematical modeling on the side, and over the course of my graduate work I transitioned to essentially the opposite. I spend most of my time doing statistical and mathematical modeling and almost none of my time out in the field or in the lab. For example, I worked on a project trying to understand the evolution of a particular type of skin cancer in a genus of freshwater fishes in Mexico. That project involved field and labwork components, as well as fitting mathematical models describing the evolution of these cancer genes to the data that we gathered.
Now I am more well known for projects related to designing surveillance systems for infectious diseases and trying to understand and disentangle the different factors behind disease emergence and persistence in populations, specifically focusing on influenza, dengue, chikungunya, whooping cough and more recently ebola.
Q: Tell us a little more about your research on epidemic surveillance and your use of Twitter data.
We are interested in developing a rigorous quantitative framework for designing, deploying and evaluating surveillance systems. Currently, it’s hard to figure out whether any proposed surveillance sites are good or bad. The problem is that these sites are chosen based on highly qualitative criteria, i.e. I just want to have a surveillance system for influenza. A better goal that would fit within a quantitative framework would be, for example, that we want to have the maximum predictive power for two-week-ahead hospitalizations associated with influenza. We probably need to be even a little more specific than that. Do we mean laboratory confirmed influenza or influenza based on a symptomatic diagnosis? Once we have such a goal, we can try to select from available data sources, like primary health care providers reporting the number of cases they see, or laboratory data. They could also include less traditional sources, such as the number of search queries associated with coughing on Google, or the number of people that tweet about having influenza.
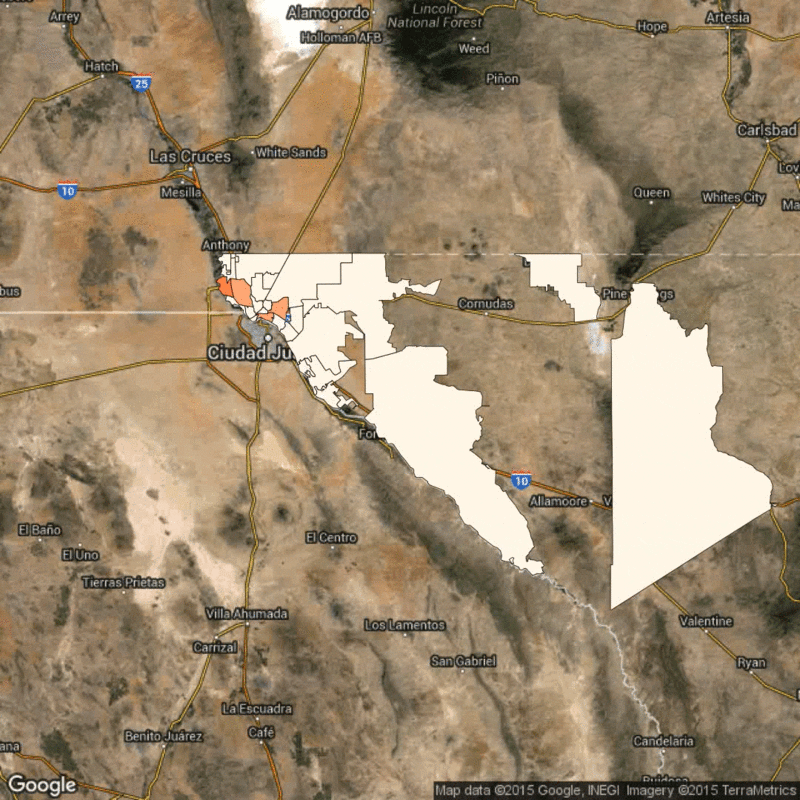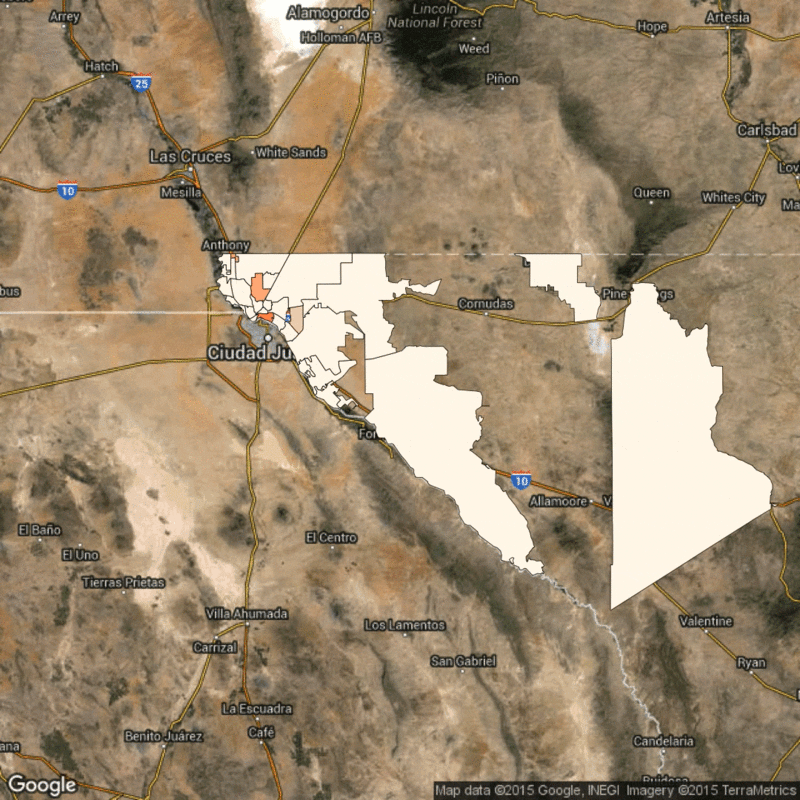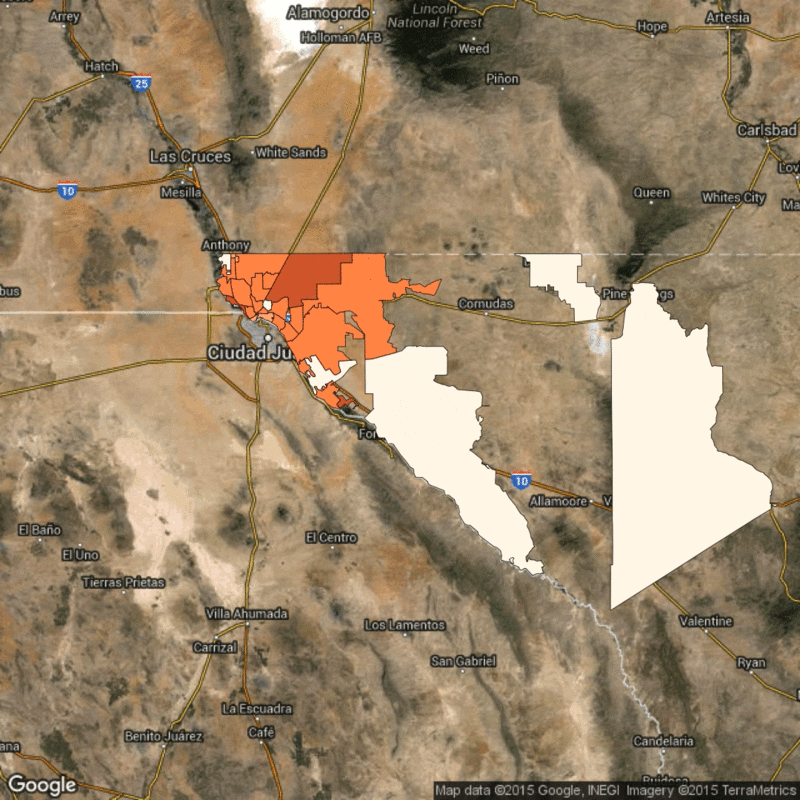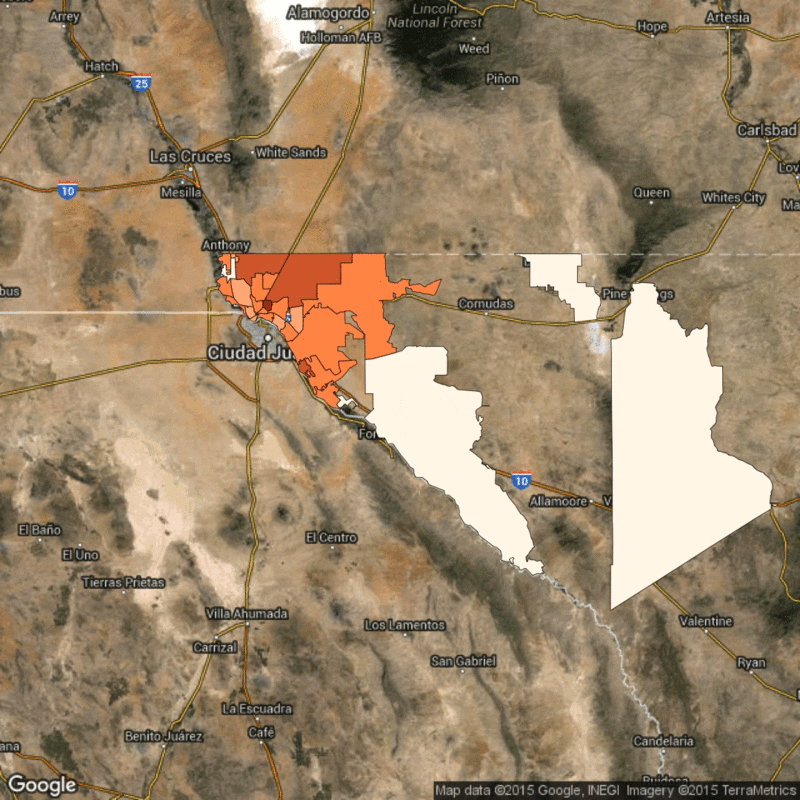
Above: Time lapse of reported flu cases in El Paso, Texas, broken down by zip code. Scarpino's research identifies how the reported flu cases in lower-income zip codes are synchronized, despite geographical distance.
More recently, we’ve also been working on using Twitter to get a lens into how some people are talking about disease and health. Obviously Twitter is a very biased sample and the types of information that people broadcast on Twitter are also biased with respect to their own true viewpoints and decision-making process. But if we can at least learn something about how people decide to wash their hands more frequently or get a vaccine or go to the doctor that will also be important for the design of future public health intervention strategies.
This idea of trying to use Twitter to learn something about thoughts and ideas surrounding taking public health action is in its infancy. Trying to use Twitter to perform surveillance is something that a number of people from a variety of different labs have been working on for years now. One of the things that has become apparent from those efforts is that people don’t tweet or search in the same way all the time. The real challenge isn’t so much that there isn’t signal of disease in Twitter or in Google or in Facebook or in the number of times people access the influenza Wikipedia page - it’s that they don’t do it in a consistent enough way that we can use it reliably, especially during a pandemic. Google flu trends failed during the 2009 flu pandemic, in part because people were searching at much higher rates relative to the number of people with actual disease because they were concerned about getting the disease.
My favorite example of this is a collaboration of mine with a researcher at Twitter to design an early warning system for Ebola. The idea we had was to look for tweets about Lassa fever, which is common in West Africa. Sometimes early Ebola cases are missed through Lassa fever misdiagnosis as both diseases can present in very similar ways to a clinician. We downloaded and looked at the time series of tweets containing the word Lassa. Sure enough, in December 2014, right when I suspected that Lassa fever tweets might spike as a result of misdiagnosis for Ebola, there was a big spike in tweets about Lassa. We got very excited and started writing the abstract of the paper. Then I got an email from my collaborator who told me to watch a particular video. It turns out there’s a band from Lassa, Turkey that released a music video in 2014 that became very popular. Once you dug into the text of the tweets, none of them were actually talking about anybody being sick - they were talking about this band from Turkey.
These types of anecdotes are very common when people try to use this type of data. There has to be an element of natural language processing where you gain an understanding of the meaning of posts, instead of just using keywords like influenza, Ebola or Lassa fever. This is part of the reason why many computer scientists and statisticians are interested in using Twitter data. It will absolutely be useful if someone can figure out how to make it work, but there are deep questions about statistics and machine learning that come up in these types of applications.
Q: How does your research relate to complex systems science?
I think the way my research most connects to complex systems science is through the many moving pieces that can influence a behavior, or an epidemic, and the interactions between those moving pieces. If we really want to understand why someone tweets a specific message, we would need to know where they were born, where they were raised, how many years of school they’ve had, how old they are, what job they have, how much disease is prevalent, how much news about the disease is prevalent, which news sources they read, which news sources they trust, whether they even use Twitter, how they use Twitter, etc. Similarly, various factors also influence whether someone walks through the door of a doctor’s office and, once there, decides whether or not to get a vaccine - that is very much a complex systems problem. You need to understand all of these moving parts and how they interact to result in one individual tweeting, and then how that individual’s tweeting can result in an epidemic of tweets, or how a sick individual can result in an epidemic of disease.
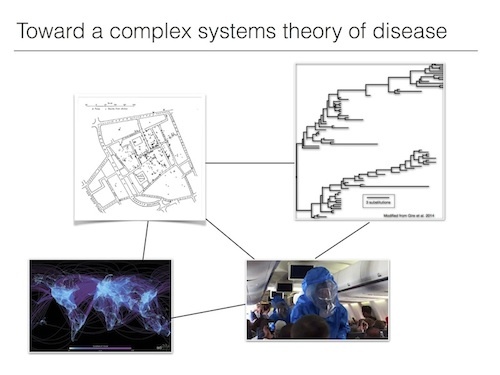
Image at right: Scarpino pieces together research on evolutionary biology, network science, human behavior and poverty to develop a more complete perspective on disease.
Another way my research connects to complex systems is that many of the hallmark processes identified in complex systems, nonlinearity, phase transitions, being out of equilibrium, are true for epidemics and most of the systems studied here at the Santa Fe Institute. For example, we are often most concerned when a disease goes from a relatively low prevalence to a relatively high prevalence; mathematically this often looks like a phase transition. Therefore, applying a complex systems approach to the study of infectious diseases allows me to leverage decades of mathematical, statistical, and computational tools to better understand epidemics. Taking such an approach is especially useful when we move beyond thinking about whether someone tweets about an illness to predicting the spread of an outbreak. Constructing such predictive models requires integrating the biology of the pathogen, the biology and behavior of the host, the evolutionary arms race between host and pathogen, and the myriad other factors all interacting to result in this one event that we care about: e.g. Ebola emerging in West Africa and killing thousands of people.
Want to hear more about Sam Scarpino’s research? Watch his recent Creative Mornings interview here.
